Abstract
Most metazoan embryos commence development with rapid, transcriptionally silent cell divisions, with genome activation delayed until the mid-blastula transition (MBT). However, a set of genes escapes global repression and gets activated before MBT. Here we describe the formation and the spatio-temporal dynamics of a pair of distinct transcription compartments, which encompasses the earliest gene expression in zebrafish. 4D imaging of pri-miR430 and zinc-finger-gene activities by a novel, native transcription imaging approach reveals transcriptional sharing of nuclear compartments, which are regulated by homologous chromosome organisation. These compartments carry the majority of nascent-RNAs and active Polymerase II, are chromatin-depleted and represent the main sites of detectable transcription before MBT. Transcription occurs during the S-phase of increasingly permissive cleavage cycles. It is proposed, that the transcription compartment is part of the regulatory architecture of embryonic nuclei and offers a transcriptionally competent environment to facilitate early escape from repression before global genome activation.
Introduction
Regulation of transcription underlies the coordination, determination and maintenance of cell identity, during organismal development. Nuclear topology and chromatin structure are key factors in the coordination of transcription of genes scattered across the genome (reviewed in ref .1). However, the relationship between spatio-temporal dynamics of transcription and the 4D organisation of the nucleus is poorly understood. Genome activation leads to the concerted activation of a large number of genes, which offers a tractable model to address the nuclear topology organisation of dynamic transcription. The earliest stages of development are under the exclusive control of maternally deposited proteins and RNAs, while the embryo remains in a transcriptionally silent state2. In externally developing metazoan embryos a series of extremely fast and metasynchronous cell division cycles precede global zygotic genome activation. Genome activation is regulated by a threshold nucleo-cytoplasmic (NC) ratio, which is reached at the mid-blastula transition (MBT)3 and reflects release from repression by diluted maternal factors, such as histones and replication factors4,5,6,7, Together with genome activation, simultaneous clearance of maternal RNAs8 at MBT overhauls the embryonic transcriptome9.
There is accumulating evidence for robust RNA Polymerase II (Pol II) transcription prior to global zygotic genome activation, in most animal models2. A small group of genes are activated several cell cycles before the MBT and represent the first wave of genome activation10,11,12. The first genes expressed in the zebrafish embryo include microRNAs, which drive the clearance of maternal mRNAs13, as well as transcription factors and chromatin binding proteins, which may play a role in the main wave of genome activation10. The existence of this first wave of genome activation raises the question of how genes escape the repressive environment before the threshold NC is reached at MBT. Furthermore, it remains unknown how transcription can occur during the short cell cycles consisting only of S and M-phases. To be able to address these questions, transcription monitoring at single-cell resolution is necessary, whilst maintaining the developing embryo context. This can only be achieved by in vivo imaging. Current imaging technologies are based on synthetic transgenic reporters of stem loop RNA-binding proteins, fused to fluorescent proteins14. This technology allowed monitoring of the dynamics, variation and nuclear topological constraints of gene expression (e.g.15,16,17,18.). However, its limitation is the requirement for transgenic manipulation of each gene of interest.
Here, native RNAs of endogenous genes were imaged, without the need to introduce fluorescent proteins by transgenesis. We developed a novel method based on arrays of fluorescently tagged antisense oligonucleotides19 for in vivo imaging of transcript accumulation and called it MOrpholino VIsualisation of Expression (MOVIE). MOVIE was used to demonstrate that the earliest gene expression in zebrafish embryos is confined to a unique transcription compartment, which forms during the S-phase of extremely short cleavage stage cell cycles, without gap phases. Nuclear organisation of transcription suggests a previously unappreciated nuclear body formation, which is seeded by transcribed loci on pairs of homologue chromosomes and persists in a cell cycle length-dependent manner. MOVIE enabled investigation of the spatio-temporal dynamics of this transcription compartment, which is a characteristic feature of pre-MBT embryos.
Results
MOVIE detects native gene transcription in living embryos
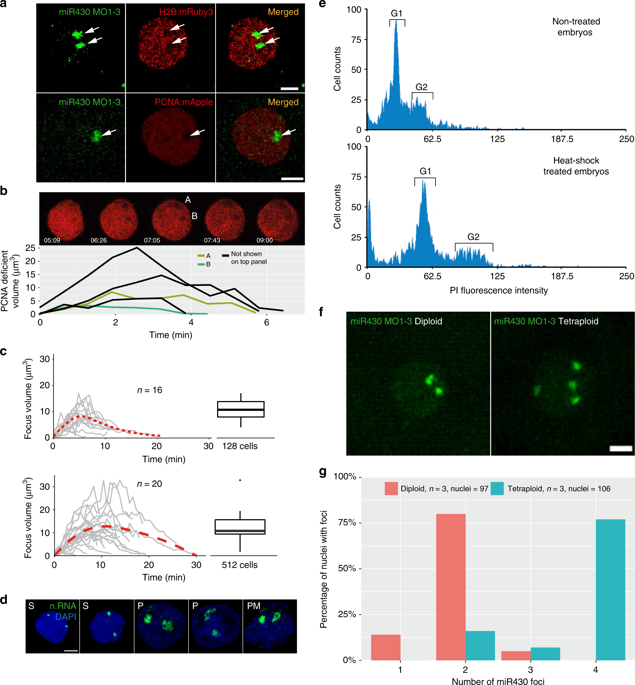
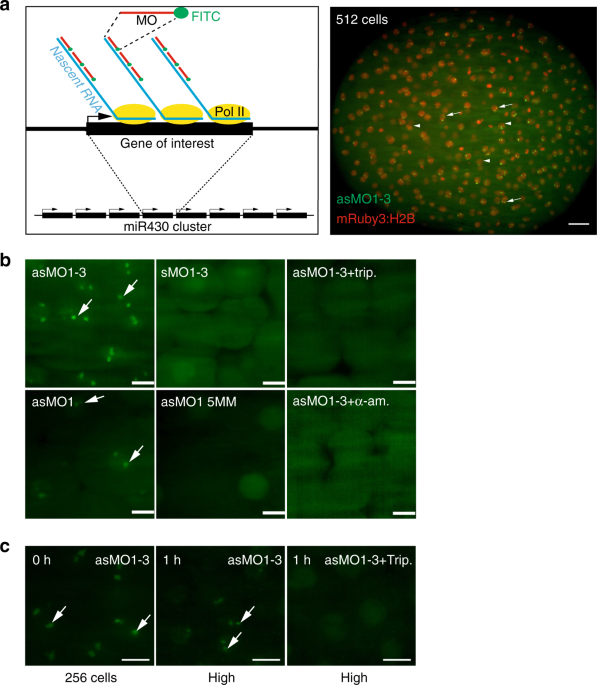
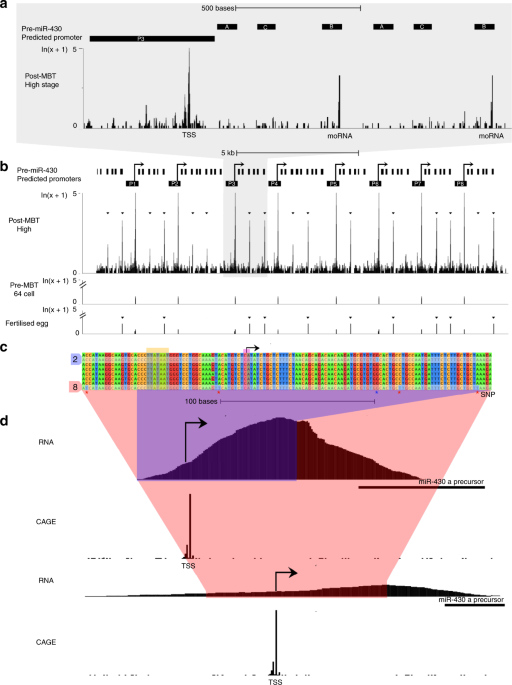
We designed a morpholino oligonucleotide (MO) array and hypothesized that binding of morpholinos at 5′ ends of transcripts will lead to detectable fluorescence signal, due to their accumulation at endogenous gene loci, in living cells (Fig. 1a). We chose the primary transcripts of miR430 microRNA genes as test for our approach. MiR430genes are the earliest and highest expressed genes during the first wave of zygotic gene activation10 and play a key role in the clearance of maternally deposited mRNA during MBT. Cap analysis of gene expression (CAGE) sequencing datasets20 were used to map the transcription start sites of the miR430 gene cluster. CAGE data revealed at least eight transcriptional start sites on the reference zebrafish genome (GRCz10), with 6-9 miR430 genes predicted to be transcribed from each of them (Fig. 2a, b). Two of these promoters contain single nucleotide polymorphisms (SNPs) allowing RNA-seq data to be uniquely mapped. This analysis demonstrated that at least two miR430promoters are used by the embryo (Fig. 2c, d). It was hypothesized that multiple promoter use at the locus may facilitate local enrichment of targeted MO fluorescence. A series of three miR430– targeting MOs (Supplementary Table 1) were generated, which upon microinjection into fertilised eggs led to the detection of two distinct transcription foci in nuclei of zebrafish blastula embryos, by lightsheet imaging (Fig. 1a, b). MO signal followed the developmental stages when miR430 expression has previously been described11,20, (Supplementary Fig. 1a). Expression signals were detected as pairs of foci per nucleus, suggesting detection of miR430 RNAs from both parental alleles. The signal was RNA dependent, as indicated by lack of signal by sense MO, was specific to miR430 RNAs demonstrated by loss of signal with mismatch-containing MO and was lost upon transcription inhibition (Fig. 1b, c). We treated embryos with triptolide after detection of robust transcription at 256 cell stage (8th cell-division) and found fluorescent foci were lost by high stage (10th cell-division), indicating that transcripts produced earlier were not detectable in subsequent stages, in line with detection of newly transcribed RNAs by MOs (Fig. 1c). Additionally, MO injection did not affect miR430 RNA production or development of the embryos (Supplementary Fig. 1b,c). We thus demonstrated detection of native and nascent miR430 primary RNA transcription in normally developing living vertebrate embryos by our novel method, which does not require transgenic manipulation.
Transcription follows cleavage stage cell cycle periodicity
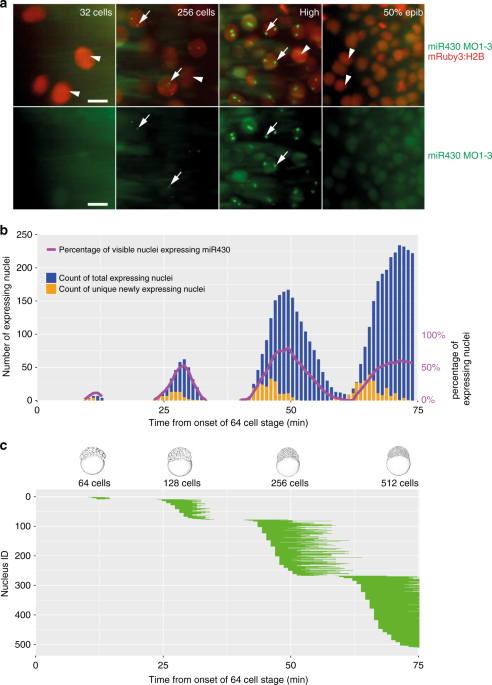
Zebrafish cleavage stages are characterised by metasynchronous cell cycles3 in which synthesis and mitosis phases are cycling without gap phases21,22,23. To understand the temporal relationship between miR430transcription and the fast cell cycles of cleavage stages, we simultaneously monitored miR430 expression by 4D time lapse imaging with MOVIE, and cell cleavages using the nuclear marker mRuby3:H2B (Fig. 3a, Supplementary Movie 1). Quantification of the miR430 transcript detection led to three main observations. Firstly, miR430expression was first detected at 64-cell stage, matching previous transcriptome analyses10,20,24, (Fig. 2a) and was present until late epiboly stages in diminishing numbers of cells. Secondly, the number of cells with miR430 expression gradually increased during cleavage stages, potentially explaining the increase in genome-wide miR430expression quantification data from whole embryos (Fig. 3b, Supplementary Fig. 1a). By the 512-cell stage all detectable nuclei show miR430 expression, however due to the loss of synchrony of this cell cycle the total number of cells expressing at any given time peaks at around 60% (Fig. 3c, Supplementary Fig. 2a). Thirdly, the temporal distribution of the start and extent of the MO signal showed gradually expanding asynchrony (Fig. 3c), which is in line with the previously described25 sequentially increasing length of pre-MBT cell cycles after the 6th cell cycle (64-cell stage). Transcriptional activity in these cell cycles also followed a spatial wave-like pattern running across the animal pole (Supplementary Movie 2). There was no obvious allelic (parental) bias observed, with most cells showing two foci of activity. Thus transcription of miR430 genes, first detected at the 64-cell stage, does not appear to respond to a precisely coordinated developmental stage timer. Instead, miR430 expression appears in sequentially increasing number of nuclei and appears to be coordinated with the overall timing of cell cycles occurring with gradually increasing asynchrony.
Transcription occurs in S-phase of elongating cell cycles
The single-cell resolution of MOVIE allows the temporal dynamics of each cell’s transcriptional activity during metasynchronous cell cycles to be addressed. Tg(Xla.Eef1a1:h2b-mRFP1) transgenic embryos26 or mRuby3:H2B microinjected protein were used to analyse cell cycle length. Neither the red fluorescence-tagged H2B, nor MO injection affected the cell cycle (Supplementary Fig. 2b,c), allowing the accurate monitoring of the relationship between cell cycle and transcription. Cell cycle length was determined by measuring the time between two anaphases. Elongation of these cell cycles, which was previously demonstrated to be due the elongation of the S-phase21 was accompanied by elongation of miR430 signal detection (Fig. 4a, b). Cell cycle length varies among cells of a single cleavage stage21, therefore correlation between miR430 expression length and cell cycle length was investigated among individual cells within a single cleavage stage. As shown in Fig. 4c, positive correlation was observed between both cell cycle length and S-phase length, but not mitosis length versus miR430expression. Next, we asked when miR430 expression occurs within the alternating M and S-phases of the pre-MBT cell cycles. By using morphology distribution and entropy of mRuby3:H2B pixels as indicators for chromatin compaction, the cell cycle was segmented into its mitotic phases (Supplementary Fig. 2d) with the length of these phases measured (Fig. 4a, d). We detected the start of miR430 activity universally during the S-phase of the pre-MBT cell cycles, but with varying delay (Fig. 4d), while the lowest levels of activity are potentially missed by the imaging approach.
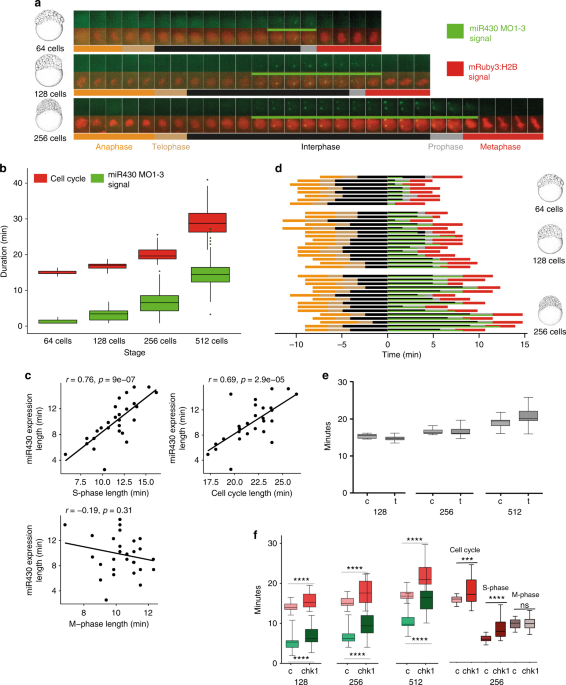
An important observation from the cell cycle phase segmentation analysis was the detection of MO signals, albeit in reduced volume and intensity, into the prophase and even extending into metaphase, either suggesting transcription during M-phase or transcript retention at the transcription site (Fig. 4a, d, Supplementary Fig. 2e).
We have then asked whether transcription activity of mother cells has impact on transcription activity of daughter cells, which potentially indicate existence of transcription memory15,27. To this effect we clustered nuclei at 128 cell stage into expressing and non-expressing groups and then tracked their daughters at 256 and 512 cell stages. There was a statistically significant increase observed in both the on/off state and length of transcript detection in daughter cells that emerged from expressing mother cells (Supplementary Fig. 2f-h).
Spatio-temporal coupling of replication and transcription at the main wave of zygotic genome activation has previously been established28,29,30, yet this has not been explored during the little-studied first wave of genome activation. Our observation that the temporal extension of miR430 activity matches that of the corresponding cell cycle lengthening, suggests the hypothesis that transcriptional activity is co-regulated with cell cycle elongation during cleavage stages. To test if transcription triggers cell cycle elongation at these early stages, transcription initiation was blocked by triptolide and its effect confirmed by the observation of loss of epiboly movements, as described previously31 (Supplementary Fig. 3a). No significant change was observed in cell cycle length, suggesting that cell cycle elongation in cleavage stage embryos is not explained by transcriptional activation (Fig. 4e). In contrast, when precocious Checkpoint kinase 1 (Chk1) activation was forced by injecting synthetic chk1 mRNA32,33, significant elongation of the cell cycle in cleavage stage embryos and corresponding elongation of miR430 MO signal were detected in every cleavage cycle measured (Fig. 4f, Supplementary Fig. 3b,c). This elongation of the cell cycle was due to changes in S-phase as no significant changes were seen during M-phase (Fig. 4f). Thus, cell cycle elongation is permissive for miR430 transcription and/or transcript retention in the sequentially expanding S-phases during cleavage stages in zebrafish.
Transcription of first wave genes in a shared compartment
The observation of a pair of pri-miR430 foci raises the question of the nature and dynamics of this compartment in relation to other transcriptional activities, during the first wave of genome activation. An antibody that recognises phosphorylated serine-2 in the C-terminal domain (CTD) of the large subunit of RNA Polymerase II (Pol II Ser2P) was used as a marker of transcription elongation34. This revealed two large foci, similar to miR430 in pre-MBT stages, that expand to broadly distributed irregular speckles after the main wave of global genome activation, likely reflecting the large expansion of transcription at thousands of genes9,11(Fig. 5a). Next, an in vivo approach was tested to simultaneously detect mir430 and Pol II SerP2 activities. A single miR430 targeting MO, previously observed to be sufficient to detect miR430 activity alone (Fig. 1b), was chosen for labelling with Cy5 (Supplementary Fig. 6), co-injected with labelled Fab fragments against Pol II Ser2P35 into embryos and imaged by confocal microscopy in vivo. Colocalisation was observed between Pol II Ser2P and miR430 MOs in live embryos, indicating that these newly described miR430-containing domains are transcriptionally highly active at pre-MBT stages (Fig. 5b) and appear to be the main site of Pol II activity at these stages. The colocalisation of Pol II SerP2 with miR430 transcripts was also verified by immunostaining (Fig. 5b). To further characterise cleavage stage transcription sites by an independent approach, a nucleotide analogue incorporation assay36 was used and nascent transcripts in pre-MBT embryos visualised with and without Pol II Ser2P localisation. This showed colocalisation of the majority of nascent RNAs together with Pol II Ser2P (Fig. 5c). Pol II Ser2P signal appeared as a dense accumulation of multiple speckles throughout the transcription compartment, marked by nascent RNA (Fig. 5c, Supplementary Fig. 5a, Supplementary Movie 7)
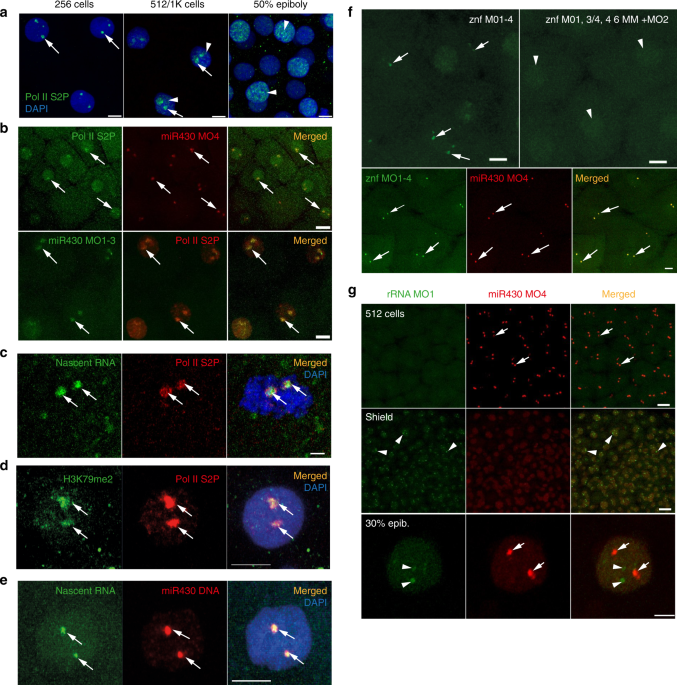
Furthermore, immunohistochemical detection of H3K79me2, an indicator of transcriptionally active chromatin37, at the site of Pol II Ser2P provides additional evidence for the majority of transcriptional activity being confined to two transcription compartments at these stages of development (Fig. 5d). A 200kbp BAC probe containing the miR430 locus was detectable in nuclei as large, but distinct nuclear territories and colocalised with the main site of nascent RNA detection (Fig. 5e). This supports the suggestion that miR430 activity marks the transcriptional compartment, which appears as the main site of early transcription and is characteristic of the first wave of genome activation.
Next, we asked whether expression of other genes, besides miR430, contribute to the formation of this transcription compartment. The expression of a partially annotated set of zinc finger (znf) genes, consisting of over 350 copies localised on the same chromosome as mir430 (chr4)11,38 and peaking at main wave of ZGA11 were investigated during the first wave of genome activation using CAGE-seq data(Supplementary Fig. 4a). From these, 25 znf genes were found to be highly conserved (Supplementary Fig. 4b), and highly active at pre-MBT and scattered across a 37 Mbp region, immediately downstream of the miR430 gene cluster on chromosome 4. The pre-MBT activity of znfgenes suggested them as candidates for transcription visualisation and we designed MO sequences to the highly conserved 5′ end of their transcripts (Supplementary Fig. 4b, Supplementary Table 1). A series of quadruple MOs, which together target all 25 znfs (Supplementary Fig. 4c) were injected into embryos and led to specific detection of znfgene activity in two foci similar to miR430 during cleavage and MBT stages (Fig. 5f). Using two-colour labelling of MOs for the simultaneous detection of fluorescein-tagged znf MOs and Cy5-tagged miR430 MO4, colocalisation of these two sets of RNAs, in a pair of shared transcription compartments, was demonstrated (Fig. 5f, Supplementary Movie 3).
Next the relationship of the observed transcription compartment with other compartment forming nuclear transcriptional activities was investigated. Four out of five loci of 45 S rDNA, which encode Pol I-transcribed ribosomal RNAs (rRNA) reside on chromosome 439. Whether nucleoli forming rRNAs can also be visualised similarly to Pol II transcribed genes on the same chromosome was queried. Using MOVIE it was found that rRNA transcripts can be detected, however only after the main wave of genome activation at MBT, with low signals at 30% epiboly and frequent activity at shield stages (Fig. 5g), which is in line with previously described rRNA expression patterns36,39). At post-MBT stages, miR430 transcripts were still detected in a mosaic fashion (Fig. 5g) and allowed subnuclear localisation analysis of both RNAs when both gene products were detected in the same cells. Thus dual MO labelling permitted miR430 RNA and rRNA distribution to be compared. At 30% epiboly stage their accumulation was distinctly localised (Fig. 5gbottom row). This result demonstrates that the transcription compartment containing Pol II gene products is independent from the nucleolus-forming RNA compartment despite 4 of 5 annotated rRNA genes reside on chromosome 4.
Transcription compartment and nuclear architecture
Our results, obtained by several imaging tools, demonstrate the formation and prominence of a major transcription compartment, characterised by localised Pol II Ser2P enrichment and high concentration of newly synthesized nascent RNAs of several genes from at least two sets of gene families. Thus, it may fulfil several of the criteria for a nuclear body (reviewed in ref .40). Among them, local depletion of chromatin may occur, which has recently seen during formation of transcriptional microenvironments during the main wave of genome activation41. We therefore analysed chromatin state in pre-MBT embryos by simultaneous MOVIE imaging of miR430 RNA, together with mRuby3 labelled H2B and mApple labelled Proliferating Nuclear Antigen (PCNA), which is broadly localised to DNA in the nucleus during S-phase of cleavage stages in zebrafish42. Reduction of both chromatin-associated proteins was observed at the site of miR430 RNA accumulation and this depletion correlated with the dynamics of RNA detection (Fig. 6a). This lack of chromatin in a compartment was also seen without MO injection, or in embryos where endogenous Pol II Ser2P was detected without MO injection (Fig. 6b). Thus, we describe a nuclear transcription compartment, with coding and non-coding Pol II transcribed nascent RNAs, large quantities of transcriptionally active Pol II and local depletion of compact chromatin, which together suggest the formation of a transcription body in early embryos.
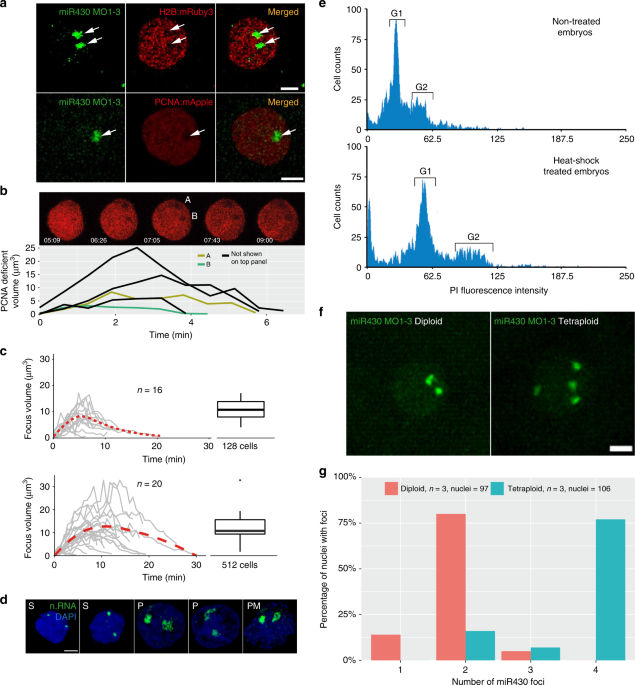
The temporal dynamics of the nuclear compartment in relation to cleavage stage cell cycles was addressed next. Measurement of miR430 MO signal and chromatin depletion volumes indicated highly dynamic formation, maintenance and disintegration of this compartment within a cell cycle (Fig. 6b, c, Supplementary Fig. 5b, Supplementary Movie 4). The transcription compartment has a characteristic volume dynamic, which shows gradual increase at both 128 and 512-cell stages peaking approximately at 16 μm3. At 512-cell stage the maximum volume shows an extended plateau in comparison to 128-cell stage, which correlates with the extended cell cycle (Fig. 6c, Supplementary Movie 5). The transcription compartment and its dynamics were also confirmed independently without MO injection, in fixed embryos, using nascent RNA detection by EU incorporation (Fig. 6d).
The presence of two transcription compartments in nuclei suggests that their formation is seeded by anti-paired43 homologue chromosomal loci occupying distinct chromosomal territories. To test the role of chromosomal organisation in forming transcription compartments, tetraploid embryos were generated by blocking centrosome separation during the first cell division using heat shock44and their ploidy verified by FACS (Fig. 6e, f). The miR430 activity foci were counted and it was found that the overall majority matched diploid allele count, whereas the number of foci was double that of diploid’s, in tetraploid embryos (Fig. 6g, Supplementary Movie 6). This finding suggests that the sites and number of transcription compartments are regulated by alleles on homologue chromosomes during cleavage stages. Interestingly, tetraploids, which have double the NC ratio and initiate MBT one cell cycle earlier than normal diploid wild-type embryos3, did not activate miR430 expression earlier than diploids, which argues against a NC ratio threshold being the trigger for their gene expression before MBT (Supplementary Fig. 5c).
Discussion
Here we describe a unique transcription compartment, the formation of which is characteristic to the first wave of gene expression in the embryo. Our results suggest that pre-MBT transcription escapes the global repressive effect of diluted maternal factors via the formation of a dynamic transcription compartment, where most detectable transcription from at least two gene families takes place. The observation that the majority of nascent RNAs and Pol II Ser2P colocalise with local depletion of chromatin, indicates the formation of a primordial nuclear body as the major site of detectable transcriptional activity, prior to global genome activation at MBT.
The temporal dynamics of this transcription compartment follows and is co-regulated with the metasynchronous cell divisions, both of which gradually elongate and increasingly lose synchrony at each cell cycle. This challenges long held views on MBT as a watershed stage; instead supporting a gradual process model for genome activation, previously proposed, based on gradually elongating cell cycles45. By manipulating transcription and cell cycle progression it was demonstrated herein that these sequentially expanding cell cycles are permissive for transcription and transcript accumulation and that the majority of transcriptional activity is during S-phase of the pre-MBT cell cycles, which lack gap phases23,46. Yet, it is unclear whether the observed presence of transcripts during prophase and prometaphase are remnants of nascent RNAs, or indicate de novo transcription. It is notable, that Pol II Ser2P is still present at prophase, potentially reflecting transcriptional activity in M-phase while it is unclear, whether this reflects elongating polymerase.
The mechanisms that allow pre-MBT transcription are likely different from that underlying the main wave of genome activation. Several mechanisms were suggested to underlie transcription activation in early embryos. These include a NC ratio threshold acting via titration of maternal factors4, a developmental timer acting via delayed access to transcriptional activators regulated by time-limited translation of maternal mRNAs (e.g9.), or availability of threshold amount of DNA template (reviewed in ref .8,47). In our experiments NC ratio does not appear to regulate the earliest of gene activation, as manipulation of nuclear content by inducing polyploidy did not have a marked effect on the timing of when expression is first detected. Furthermore, our observations on pre-MBT transcription detection at the single-cell level and with specific loci also exclude the model, which argues for lack of detectable transcription due to limiting amount of DNA template. Instead, our data suggests alternative mechanisms, which could include delayed availability of transcriptional activators, for example by the control of maternal mRNA translation.
The formation of the nuclear body may occur by several mechanisms. It is possible that the unique, repetitive organisation of the long arm of chromosome 411 which includes the miR430 cluster, as well as a series of znf genes together creates a local highly transcribing environment, which may function as an aggregation of transcription factories48,49 and appears as a distinguishable body. Interestingly, the miR430 locus was shown to be specifically enriched for Rad21 binding before zygotic genome activation50, which suggests the involvement of cohesins in the formation of a distinct local chromatin environment. Additional contributing factors to the formation of the large compartment may involve phase separation principles40,41 or other means of local accumulation of RNA at the transcription site. Nevertheless, by detection of specific chromosomal loci and their RNA products, as well as by ploidy manipulation, we demonstrated that these nuclear bodies are seeded by DNA and that the candidate loci for coordinating these nuclear compartments reside on homologues of chromosome 4 occupying distinct territories in the nucleus. While our rRNA imaging data suggests separation of Pol II and nucleolus-forming rRNA compartments, future studies will be necessary to investigate the relationship between the transcription compartment described here and other nuclear bodies, such as Cajal body and the histone locus body36 during early zebrafish development.
The formation of the transcription compartments may serve several purposes in the uniquely fast cell division cycles that precede global genome activation. Firstly, it may allow for efficient transcription by concentrating transcription apparatus into a confined nuclear space, at a time when activators and transcription machinery components are likely rate limiting due to delayed translation of maternal stockpiles of their mRNAs24,51. Secondly, restricted localisation of transcription into a compartment may indicate an as yet unexplored mechanism for facilitating the escape from the general repressive environment of the nucleus, which is only removed from the rest of the genome by reaching a certain NC ratio. Initial observations however, suggest that the first wave of transcription is not affected by similar titration of maternal repressive factors as the rest of the genome, as indicated by unaffected initiation of transcription in tetraploids, where the NC ratio is doubled. Thirdly, it may provide spatial and temporal separation of replication and transcription—two potentially conflicting genome activities occurring in the short S-phases without gap52. The transcription compartment with activities of genes residing on chromosome 4 raises the question of where first wave-expressed genes residing on other chromosomes are transcribed in the pre-MBT nucleus. While most Pol II Ser2 and nascent RNAs are detected to colocalise with the pri-miR430 signal, it is not excluded that low expressing loci are active in a separate domains from the chromosome 4 territory. Future work aiming to improve detection of low expressed single copy gene products will provide insight into the generality of the transcription compartment described in this study.
The compartmentalisation of the first wave of genome activation is likely a general phenomenon and not unique to zebrafish. In situ detection of large Pol II Ser2P and Pol II pSer5 punctae in Drosophilaembryos during and before global genome activation, has been documented53,54,55,56 and may explain the first wave of genome activation not only in Drosophila, but more generally among externally developing animal models. Due to the simplicity of the MOVIE approach, testing this hypothesis in early embryos of other animal models will be feasible. Utilising multicopy gene product detection, MOVIE will be particularly useful to study RNA accumulation in nuclear bodies. Taken together, our in vivo monitoring of nuclear architecture organisation into transcription compartments contributes to the growing body of evidence supporting the notion of suprachromosomal organisation in the nucleus, serving not only structural, but also gene regulatory function57,58 and highlights the early embryo as a tractable model system to study the interface between transcription and nuclear organisation.
Methods
Characterisation of miR430 locus
Identification of predicted multiple promoter sites at the miR430 locus were determined from CAGE-seq data20 mapped to zebrafish assembly GRCz10 by Tophat59 allowing up to 80 multi-mapped reads. Start sites of transcription were determined by usage of CAGEr software60. Annotation of promoter regions (Fig. 2) were defined as 400 bases upstream and 100 bases downstream of the detected TSS with pre-miR430 gene annotations determined from Ensembl database with addition of 2 pre-miR430 c-subtype sequences found by BLAT search, completing the a-c-b triplet structure61.
Detection of unique SNPs in miR430 promoter sequences was achieved by multiple sequence alignment with Clustal Omega62. Thus allowing detection of nascent RNA10 reads aligning to two individual promoters in the miR430 cluster by unique mapping with STAR aligner63. Expression of miR430 during development is demonstrated both by CAGE-seq20 and RNA-seq11 spanning 19 developmental time points, capturing expression both pre and post-MBT (Supplementary Fig. 1a).
Morpholino design for ZNF genes on chromosome 4
Targeting MOs were designed specifically to visualise transcription of zinc finger genes on chromosome 4, most highly expressed in the early zygote, as determined from CAGE-seq dataset20. Twenty three such genes with a fully annotated transcript were identified. The 5′ UTRs of these genes with 50 bp up- and downstream flanking sequence were used for conservation assessment, via multiple sequence alignment using MUSCLE alignment algorithm64 and subsequent visualizations of inherent similarity were achieved with JalView65. Consensus sequences were generated and used for morpholino design, targeting conserved regions in the 5′UTRs. Altogether 4 morpholino were designed (Supplementary Fig. 4b).
To identify all potential znf genes on chromosome 4, targeted by the designed MOs, a BLAST search against Ensembl version 92 cDNA database was performed. A transcript was considered as a potential target by a morpholino if at least a 20 bp match with 84% identity was found. Intersections were calculated from the identified potential target genes list for each morpholino by VennMaster66 and quad venn-diagram graphic (Supplementary Fig. 4c) was generated with VennDiagram CRAN R package67.
Synthesis of Cy5-tagged miR430 MO4 morpholino
Unless otherwise stated materials and reagent were used as supplied by Sigma-Aldrich and Fisher Scientific. Morpholino sequence (I) bearing 3′-alkyne functionality attached by propyl acetate linker were obtained from Gene Tools Ltd and used as supplied. Desalting was conducted using NAP-10 columns (GE Healthcare). HPLC analysis and purification was carried out using a Shimadzu LC2010 with a Phenomenex Kinetex 5 µm C18 100 Å (250 × 4.6 mm) column. Mass spectrometry was conducted on a SYNAPT G2-S instrument using TOF MS ES + with a solvent mixture of 50:50 water/acetonitrile containing formic acid (0.1% v/v). UV absorption spectroscopy was conducted on Shimadzu UV-180 UV spectrophotometer. Solvents used were of HPLC grade, Milli-Q water is ultrapure water supplied by Millipore Corporation.
An alkyne-tagged miR430 MO4 sequence (I) was exposed to azide-tagged Cy5-fluorophore II under copper-catalysed triazole-forming conditions to deliver Cy5-tagged sequence III. Fluorescein-tagged morpholino sequence IV (and alkyne-tagged sequence I) were supplied by Gene Tools Ltd. The conversion to III is detailed in Supplementary Fig. 6a.
Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) (11 mg, 25.5 µmol), sodium abscorbate (5.3 mg, 27 µmol) copper sulphate (0.94 mg, 3.75 µmol) and a magnetic stirring bead were placed in a 2 mL vial. 3′-Alkyne morpholino sequence (I, 150 nmol, from Gene Tools) was dissolved in water (500 µL, milli-Q) and was transferred to the reaction vial. Cy5 azide (II, 225 nmol) dissolved in water (250 µL, milli-Q) was added to the reaction mixture. A further addition of water (250 µL, milli-Q) brought the total volume of the mixture to 1 mL. The mixture was stirred at room temperature for 16 h. The reaction mixture was desalted by passing through a NAP column and rinsed with additional water (0.5 mL, milli- Q) to obtained 1.5 mL of product-containing mixture.
The Cy5-tagged oligo miR430 MO4 was purified by analytical scale HPLC using conditions as follows:
Phenomenex Kinetex 5 µm C18 100 Å (250 × 4.6 mm) as the stationary phase and a defined mixture of water and acetonitrile as the mobile phase.
Flow rate: 2 mL/min
Column temperature: 35 °C
Gradient:
- (i) 95:5 water:acetonitrile (0 - > 3.5 min)
- (ii) 70:30 water:acetonitrile (3.5 - > 8.5 min)
- (iii) 0:100 water:acetonitrile (8.5 - > 13 min)
- (iv) 95:5 water:acetonitrile (0 - > 3.5 min)
Detection: UV/vis: 254, 260, 647 and 663 nm; Excitation 647 nm Emission 633 nm
Collection: A peak in the chromatogram centred at 6.1 min was collected. The peak thus described displayed absorbances at the four detection wavelengths and fluorescence under the excitation and emission conditions described. The material collected was freeze dried and re-dissolved water (100 µL, milli-Q). Concentration: The concentration was determined by UV-Vis absorption, using absorption equation A = ε.l.c(where A = absorption, l is the path length, c = concentration and the extinction coefficient (ε) of the Cy5 ε650 = 250000 M−1cm−1). The collected fraction was diluted (20 μL of sample fraction and 140 μL of water) to 160 μL of analyte solution with an observed absorption of 0.81084. Path length was 1 cm. The calculated concentration was thus determined to be 25.9 μM. The resulting solution was analysed for purity by analytical HPLC, using a Phenomenex Kinetex 5 µm C18 100 Å (250 × 4.6 mm).
Flow rate: 1 mL/min
Column temperature: 35 °C
Gradient:
- (i) 95:5 water:acetonitrile (0 - > 25 min)
- (ii) 82:18 water:acetonitrile (25 - > 30 min)
- (iii) 0:100 water:acetonitrile (30 - > 40 min)
- (iv) 95:5 water:acetonitrile (40 - > 45 min)
Detection: UV/vis 260 and 645 nm
A single peak in the chromatogram centered at 29 mins displayed absorbance at both 260 and 645 nm (Supplementary Fig. 6b,c) and had the correct m/z ([M + ] III) of 9258 by mass spectrometry (Supplementary Fig. 6d,e,f).
Antibody Fab fragments production and labelling
The mouse monoclonal antibody against human RPB1 CTD, phosphorylated at serine-2 (MABI0602) was purchased from MBL laboratories (Nagoya, Japan). Fab fragments were prepared by digesting the antibodies with papain. A total amount of 200 μg of antibodies were reduced by adding 1 mM Tris(2-carboxyethyl)phosphine (C4706, Sigma-Aldrich, St. Louis, MO) and afterwards digested using 100 μl of magnetic beads coated with papain (PAPM-40-2, Spherotech Inc., Libertyville, IL) for 3 h at 37 °C. The Fab fragments were subsequently separated from Fc fragments by purification on 100 μl of Protein A Sepharose beads (GE Healthcare, Issaquah, WA) for 30 min at 4 °C. Afterwards, 100 μg of Fab fragments were fluorescently labelled using an Alexa Fluor 568 Monoclonal Antibody Labelling Kit (A20184, Thermo Fisher, Waltham, MA) following the manufacturers protocol.
Zebrafish maintenance
All zebrafish strains were maintained in designated facility (according to UK Home Office regulations) in a recirculating system (ZebTEC, Tecniplast) at 26 °C in a 10-hour dark, 14-hour light photoperiod and fed three times daily.
Zebrafish experiments were restricted to early embryos and adults were only used for natural breeding. Animal work presented in this study was carried out under the project licences 40/3681 and P51AB7F76 assigned to the University of Birmingham, UK
Microinjection
Single-cell stage embryos were injected with 2 nl solutions containing targeting or mismatch control morpholinos at a concentration of 7 µM each in nuclease-free water, supplemented with 0.1% phenol red. Unlabelled and fluorescein-labelled morpholinos were ordered from Gene Tools LLC.
At the low concentration used, MOs did not cause developmental defects in the majority of embryos, and did not affect mature miRNA processing from the primary transcripts (Supplementary Fig. 1b,c). Upon co-injection with mRuby3:H2B, mApple:PCNA fusion proteins or antibody, the final concentration of proteins in the injection solution was 400 ng/µl for the fusion proteins and 250 ng/µl for the antibody. We verified that neither recombinant H2B protein nor injection of MO affected the cell cycle (c,c). After injection the embryos were kept in E3 media at 28 °C until imaging or fixation.
Transcription block
Transcription block was performed by treatment with 1 µM triptolide from the single-cell stage (Sigma T3652) in E3 media, or by microinjection of 200 pg α-amanitin (Sigma A2263) at single-cell stage.
Live embryo imaging
Imaging of live embryos injected with fluorescent labelled MOs, fluorescent protein fusions or antibodies was performed on Zeiss Lightsheet Z1 or Zeiss 880 confocal microscope with Fast Airyscan Module.
For imaging on Zeiss Lightsheet Z1, embryos were mounted in 1% low melt agarose column using size three glass capillaries and incubated in E3 media at 28 °C during imaging. Z-stacks of ~200–300 slices in 0.5–1 µm steps were acquired every 30–60 seconds for 1.5–2.5 h, with ×20 objective.
For imaging on the Zeiss 880 confocal microscope in Fast Airyscan mode, embryos were mounted in 0.5% low melting agarose in glass bottom imaging dish and incubation chamber temperature was maintained at 28 °C during imaging. Z-stacks of ~100–200 slices were acquired (step size of 0.25 µm) every 30–60 seconds for 1.5–2.5 h using a ×25 or ×40 objectives. Post-processing of the acquired images was performed using Zeiss ZEN Blue Software.
Analysis of spatio-temporal dynamics of MO signal
For global temporal analyses, lightsheet microscopy (morpholino/transgenic mRuby3:H2B) datasets were transformed into maximum intensity projections (MIPs), using Zeiss Zen Black software. Each MIP was imported into Icy Bioimage (versions 1.9.5.0–1.9.7.0, Quantitative Image Analysis Unit, Pasteur Institute, France), where the spot detection plugin was used to detect transcription foci and nuclei, as these features are represented as regions of locally elevated intensity, in their, respective, channels. The plugin was guided to detect features as bright regions against a darker background. Transcription foci were typically detected using a pixel size parameter of 3px, with a sensitivity of between 80 and 150% depending on noise (with higher sensitivity for higher noise). Feature size limits for detecting transcription foci were between 3 and 75 pixels in area. Nuclei were detected using a pixel size of between 13 and 25px, with similar sensitivity settings. Feature size limits for detecting nuclei were between 200 and 3000 pixels in area. Spot detection produced 2D regions of interest (ROIs) around features for each channel separately. Transcription foci were given a label which associated with their corresponding nucleus, and this was applied on a frame by frame basis to allow counting of nuclei expressing transcription foci. ROI metadata was exported to Microsoft Excel where transcription foci could be quantified.
3D analysis
3D datasets (Airyscan morpholino/injected mRuby3:H2B protein) collected from the Zeiss 880 confocal microscope with Fast Airyscan Module were imported into Icy Bioimage, where the spot detection plugin was used to identify nuclei and transcription foci, using the same sensitivity settings as described previously. Size features were scaled up to account for volumes: feature size settings for transcription foci were between 5 and 150 pixels, and size settings for nuclei were between 750 and 20,000 pixels. This would produce a 3D ROI around the detected feature. In the case of nuclei this ROI could be viewed in the 3D VTK viewer on Icy Bioimage, where a video of the rendered ROI could be recorded using the built-in recording function (Supplementary Fig. 5). ROI data were exported to Microsoft Excel where transcription foci volumes were extracted per frame and timeseries could be plotted (Fig. 6c).
In PCNA imaging data obtained on Zeiss 880 confocal with Airyscan, PCNA deficient regions were quantified using the spot detection plugin, with the setting to search for dark features against a bright background. Pixel sensitivity and 3D size settings were the same as for transcription foci detection, described previously. Relevant ROI’s were confirmed by visually inspecting that they correlated with signal deficient regions, and labelling the same feature with the same name per frame. The volume for each feature was extracted and plotted over time using Microsoft Excel.
Cell cycle segmentation reference
Cell cycle phase segmentation was based on a visual library of cell cycle phase features generated from previously published confocal imaging of chromatin68,69,70. This reference library was used as a base for segmenting nuclei from MIP Zeiss Lightsheet data into cell cycle phases. Once phases were assigned to an initial batch of nuclei, the imaging parameters were extracted. These parameters were used to generate a quantitative measure of chromatin consistency throughout the cell cycle and assign a measure of cell cycle phase to each nucleus. Imaging parameters included 2D area covered by a nucleus ROI and standard deviation in pixel intensities across the nucleus (Supplementary Fig. 2d).
Whole mount antibody staining
Embryos were fixed at the desired stage in 4% PFA in PBS then permeabilised with PBS-0.3% TritonX-100. Blocking was performed with room temperature blocking solution (BlockAid™, B10710, Thermo Fisher) then transferred to blocking solution containing primary antibody. Excess primary antibody was removed with PBS-0.1% Tween-20 washes. Samples were then incubated with blocking solution containing appropriate secondary antibody conjugated to Alexa Fluor fluorescent dye at 1:500 dilution. Immunostained embryos were imaged in antifade mounting media with DAPI (Vectashield, H-1200, Vector Laboratories) on Zeiss 880 confocal microscope with Fast Airyscan Module.
Primary antibodies used:
- Anti-RNA polymerase II (phospho S2), ab5095, Abcam at 1:400 dilution
- Pol II S2P monoclonal antibody, C15200005, Diagenode at 1:1000 dilution
- Anti-Histone H3 (di methyl K79) antibody, ab3594, Abcam at 1:1000 dilution
Nascent RNA staining
Zebrafish embryos were injected with 1 nl of 50 mM ethynyl-uridine (EU, Thermo Fisher, C10329) solution at single-cell stage, incubated in E3 media at 28 °C until fixation at the desired stage. Embryos were permeabilised with PBS-0.3% TritonX-100 before undergoing detection of nascent RNA using the Click-iT™ RNA Alexa Fluor™ 488 Imaging Kit (Thermo Fisher, C10329), following the manufacturer’s protocol. After the reaction embryos were imaged on Zeiss 880 confocal microscope with Fast Airyscan, or used for subsequent protein or DNA detection.
DNA FISH
Probes for DNA-FISH were prepared using the FISH Tag™ DNA Multicolor Kit (Thermo Fisher) following kit instructions. The BAC DKEY-69C19 was used as a template for DNA-FISH probe production, corresponding to 214 kb of the mir430 locus on chromosome 4.
Embryos were fixed at the developmental stage of interest using 4% PFA. Animal caps were isolated and a series of gradient washes was used to equilibrate the samples with hybridization buffer [50% formamide, 4x SSC, 100 mM NaPO4 pH 7.0, 0.1% Tween-20]. Embryonic DNA was denatured by incubation of samples at 70 °C for 15 min before application of probe and incubation overnight at 37 °C. Unbound probe was washed from the samples using a hybridization buffer: PBS-T gradient. Samples were counterstained with DAPI prior to imaging on a Zeiss LSM 880 with Fast Airyscan, with a 100 × 1.48 numerical aperture objective lens. 50–100 optical sections (130 nm thickness) were acquired of each nucleus. Acquired images were processed using Zen Black software (Zeiss).
Transgenic lines
The transgenic line Tg(Xla.Eef1a1:h2b-mRFP1) (http://zfin.org/ZDB-TGCONSTRCT-130711-126) used in this study was kindly provided by Sarah Baxendale (University of Sheffield, UK).
Protein synthesis
mRuby3:H2B and mApple:PCNA were PCR amplified form plasmids pKanCMV-mRuby3-10aa-H2B (Addgene, 74258) and mApple-PCNA-19-NLS-4 (Addgene, 54937), respectively, and sub-cloned into NdeI/BamHI linearized pET-28a + plasmid (in frame with the N-terminal 6xHis-Tag), using In-Fusion® HD Cloning Plus Kit (Clontech, 638910) following the manufacture instructions.
The plasmids were transformed into E. coli BL21 (DE3) and the proteins were expressed in LB media by IPTG induction at 20°C. The cell lysates containing mApple:PCNA were prepared by re-suspending in 50 mM Tris HCl; pH 8, 250 mM NaCl, 10 mM imidazole, 2 mM MgCl2 and 10% glycerol whereas mRuby3:H2B was resuspended in 50 mM Tris HCl; pH 8, 500 mM NaCl, 10 mM imidazole, 2 m M MgCl2, 5% glycerol and 0.3% Brij. All buffers were additionally supplemented with BitNuclease, PMSF and protease inhibitors. Cell lysis was improved by subjecting the lysates to sonication followed by centrifugation to clarify the cell extracts. The proteins were then purified by Nickel affinity chromatography (Generon). The proteins were dialysed against 500 mM KCl, 20 mM PIPES, 100 μM EGTA, pH6.8 using 12 kDa columns (Pur-A-Lyzer; Sigma).
Manipulation of embryonic cell ploidy by heat-shock
Tetraploid embryos were generated by heat-shock treatment, following the HS2 protocol adapted from44. Embryos were collected 2 min post fertilization and maintained in E3 media for a further 20 min at 28 °C, during which microinjection of MOs was performed. Embryos were heat-shocked between 20 and 23 min post fertilization by incubation in E3 media at 42 °C for 2 min, then returned to E3 media at 28 °C and allowed to develop normally. Embryos were monitored at the normal point of the second cell division (1 h post fertilisation (hpf)) and heat-shock treated embryos that failed to undergo cytokinesis were selected for downstream analysis, along with matched non-heat-shock treated controls. For the assessment of ploidy manipulation, embryos were collected at the 12-somite stage, dissociated with PBS-based enzyme-free cell dissociation buffer (Gibco), washed with PBS and subjected to propidium iodide DNA content analysis following manufacturer’s conditions (Invitrogen). Flow cytometry was performed on the CyAn ADP machine (Beckman Coulter) using the Summit 4.4 software package using the standard gating strategy.
Cell cycle length and gene expression experiments
Wild type embryos were injected with mRuby3:H2B protein at the single-cell stage and incubated in E3 media supplemented with 1 µM triptolide (Sigma, T3652) or vehicle control following injection. In cell cycle elongation experiments, wild-type embryos were injected at the single-cell stage with a cocktail of 200 pg of Xenopus chk1 mRNA, 200 pg miR430 MO and mRuby3:H2B protein whereas controls were injected with 200 pg mCherry mRNA + 200 pg miR430 MO and mRuby3:H2B protein. Embryos were imaged using Zeiss Lightsheet Z.1 microscope with the ZEN software. Images were analysed and timing of cell cycle transition from anaphase to anaphase were identified by mRuby3:H2B pattern variation using Icy Bioimage. Statistical analysis was carried out by Prism GraphPad 5 (USA). Chk1 and mCherry mRNAs were produced from NotI linearized pCS2 + :chk1 (kindly provided by Phil Zegerman) and pCS2 + :mCherry vectors, respectively, using mMESSAGE mMACHINE SP6 Transcription Kit (ThermoFisher Scientific).
Quantification of miRNA levels by digital droplet PCR
Total RNA was extracted from a 100 miR430 targeting MO1-3 and non-injected embryos at sphere stage, using TRIzol™ Reagent (ThermoFisher, 15596026) following manufacturer’s instructions. Three RNA samples for each group (MOs injected and non-injected) from three independent experiments were prepared. cDNA was synthesized from equal amounts of total RNA (100 ng) using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer’s protocol. cDNA samples were diluted 1:30 with nuclease-free water. qPCR reactions were prepared using the ddSupermix for Probes (Bio-rad) and Taqman miRNA probes miR430a (004365_mat) and miR430b (465767_mat). Droplet generation oil was added to the qPCR mixture and droplets were generated with the QX100 droplet generator (Bio-rad). PCR protocol as per manufacturer (Taqman miRNA assay) and the samples were run in a QX100 droplet reader. Quantasoft software (Bio-rad) was used for data acquisition and analysis.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The source data underlying Figs. 3b,c; 4b–f; 6b,c,g and Supplementary Fig. 1a-c; 2a-d,f-g; 3b,c; 4c; 5b are provided as a Source Data File. All 3D, 4D imaging data are available upon request. A reporting summary for this Article is available as a Supplementary Information file.
Additional information
Journal peer review information: Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bickmore, W. A. The spatial organization of the human genome. Annu Rev. Genom. Hum. Genet 14, 67–84 (2013).
- Show contextfor reference 1
- 2.Tadros, W. & Lipshitz, H. D. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042 (2009).
- Show contextfor reference 2
- 3.Kane, D. A. & Kimmel, C. B. The zebrafish midblastula transition. Development 119, 447–456 (1993).
- Show contextfor reference 3
- 4.Newport, J. & Kirschner, M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 30, 687–696 (1982).
- Show contextfor reference 4
- 5.Collart, C., Allen, G. E., Bradshaw, C. R., Smith, J. C. & Zegerman, P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 341, 893–896 (2013).
- Show contextfor reference 5
- 6.Joseph, S. R. et al. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife 6, e23326 (2017).
- Show contextfor reference 6
- 7.Amodeo, A. A., Jukam, D., Straight, A. F. & Skotheim, J. M. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl Acad. Sci. USA 112, E1086–E1095 (2015).
- Show contextfor reference 7
- 8.Lee, M. T., Bonneau, A. R. & Giraldez, A. J. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev. Cell Dev. Biol. 30, 581–613 (2014).
- Show contextfor reference 8
- 9.Harvey, S. A. et al. Identification of the zebrafish maternal and paternal transcriptomes. Development 140, 2703–2710 (2013).
- Show contextfor reference 9
- 10.Heyn, P. et al. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 6, 285–292 (2014).
- Show contextfor reference 10
- 11.White, R. J. et al. A high-resolution mRNA expression time course of embryonic development in zebrafish. Elife 6, e30860 (2017).
- Show contextfor reference 11
- 12.Chen, K. et al. A global change in RNA polymerase II pausing during the drosophila midblastula transition. Elife 2, e00861 (2013).
- Show contextfor reference 12
- 13.Giraldez, A. J. et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 (2006).
- Show contextfor reference 13
- 14.Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998).
- Show contextfor reference 14
- 15.Ferraro, T. et al. Transcriptional memory in the drosophila embryo. Curr. Biol. 26, 212–218 (2016).
- Show contextfor reference 15
- 16.Campbell, P. D., Chao, J. A., Singer, R. H. & Marlow, F. L. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development 142, 1368–1374 (2015).
- Show contextfor reference 16
- 17.Darzacq, X. et al. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 14, 796–806 (2007).
- Show contextfor reference 17
- 18.Chubb, J. R., Trcek, T., Shenoy, S. M. & Singer, R. H. Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006).
- Show contextfor reference 18
- 19.Li, W. M., Chan, C. M., Miller, A. L. & Lee, C. H. Dual functional roles of molecular beacon as a MicroRNA detector and inhibitor. J. Biol. Chem. 292, 3568–3580 (2017).
- Show contextfor reference 19
- 20.Nepal, C. et al. Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res 23, 1938–1950 (2013).
- Show contextfor reference 20
- 21.Olivier, N. et al. Cell lineage reconstruction of early zebrafish embryos using label-free nonlinear microscopy. Science 329, 967–971 (2010).
- Show contextfor reference 21
- 22.Siefert, J. C., Clowdus, E. A. & Sansam, C. L. Cell cycle control in the early embryonic development of aquatic animal species. Comp. Biochem Physiol. C. Toxicol. Pharmacol. 178, 8–15 (2015).
- Show contextfor reference 22
- 23.Zamir, E., Kam, Z. & Yarden, A. Transcription-dependent induction of G1 phase during the zebra fish midblastula transition. Mol. Cell Biol. 17, 529–536 (1997).
- Show contextfor reference 23
- 24.Lee, M. T. et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature503, 360–364 (2013).
- Show contextfor reference 24
- 25.Olivier, N. et al. Cell Lineage Reconstruction of Early. Science 70, 967–971 (2010).
- Show contextfor reference 25
- 26.Rodriguez-Aznar, E., Barrallo-Gimeno, A. & Nieto, M. A. Scratch2 prevents cell cycle re-entry by repressing miR-25 in postmitotic primary neurons. J. Neurosci. 33, 5095–5105 (2013).
- Show contextfor reference 26
- 27.Muramoto, T., Muller, I., Thomas, G., Melvin, A. & Chubb, J. R. Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr. Biol. 20, 397–406 (2010).
- Show contextfor reference 27
- 28.Schubeler, D. et al. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet 32, 438–442 (2002).
- Show contextfor reference 28
- 29.Pourkarimi, E., Bellush, J. M. & Whitehouse, I. Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. Elife 5, e21728 (2016).
- Show contextfor reference 29
- 30.Blythe, S. A. & Wieschaus, E. F. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell160, 1169–1181 (2015).
- Show contextfor reference 30
- 31.Muller, F., Lakatos, L., Dantonel, J., Strahle, U. & Tora, L. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol. 11, 282–287 (2001).
- Show contextfor reference 31
- 32.Collart, C., Smith, J. C. & Zegerman, P. Chk1 Inhibition of the Replication Factor Drf1 Guarantees Cell-Cycle Elongation at the Xenopus laevis Mid-blastula Transition. Dev. Cell 42, 82–96 e3 (2017).
- Show contextfor reference 32
- 33.Zhang, M., Kothari, P., Mullins, M. & Lampson, M. A. Regulation of zygotic genome activation and DNA damage checkpoint acquisition at the mid-blastula transition. Cell Cycle 13, 3828–3838 (2014).
- Show contextfor reference 33
- 34.Brookes, E. & Pombo, A. Code breaking: the RNAPII modification code in pluripotency. Cell Cycle 11, 1267–1268 (2012).
- Show contextfor reference 34
- 35.Conic, S. et al. Imaging of native transcription factors and histone phosphorylation at high resolution in live cells. J. Cell Biol. 217, 1537–1552 (2018).
- Show contextfor reference 35
- 36.Heyn, P., Salmonowicz, H., Rodenfels, J. & Neugebauer, K. M. Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos. RNA Biol. 14, 752–760 (2017).
- Show contextfor reference 36
- 37.Schubeler, D. et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 (2004).
- Show contextfor reference 37
- 38.Howe, K. et al. Structure and evolutionary history of a large family of NLR proteins in the zebrafish. Open Biol. 6, 160009 (2016).
- Show contextfor reference 38
- 39.Locati, M. D. et al. Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA23, 1188–1199 (2017).
- Show contextfor reference 39
- 40.Mao, Y. S., Zhang, B. & Spector, D. L. Biogenesis and function of nuclear bodies. Trends Genet 27, 295–306 (2011).
- Show contextfor reference 40
- 41.Hilbert, L. et al. Transcription establishes microenvironments that organize euchromatin. Preprint at https://www.biorxiv.org/content/early/2017/12/15/234112(2017).
- Show contextfor reference 41
- 42.Zhang, M., Kothari, P. & Lampson, M. A. Spindle assembly checkpoint acquisition at the mid-blastula transition. PLoS One10, e0119285 (2015).
- Show contextfor reference 42
- 43.Joyce, E. F., Erceg, J. & Wu, C. T. Pairing and anti-pairing: a balancing act in the diploid genome. Curr. Opin. Genet Dev. 37, 119–128 (2016).
- Show contextfor reference 43
- 44.Baars, D. L., Takle, K. A., Heier, J. & Pelegri, F. Ploidy Manipulation of Zebrafish Embryos with Heat Shock 2 Treatment. J Vis Exp(2016): 54492
- Show contextfor reference 44
- 45.Farrell, J. A. & O’Farrell, P. H. From egg to gastrula: how the cell cycle is remodeled during the Drosophila mid-blastula transition. Annu Rev. Genet 48, 269–294 (2014).
- Show contextfor reference 45
- 46.Dalle Nogare, D. E., Pauerstein, P. T. & Lane, M. E. G2 acquisition by transcription-independent mechanism at the zebrafish midblastula transition. Dev. Biol. 326, 131–142 (2009).
- Show contextfor reference 46
- 47.Langley, A. R., Smith, J. C., Stemple, D. L. & Harvey, S. A. New insights into the maternal to zygotic transition. Development 141, 3834–3841 (2014).
- Show contextfor reference 47
- 48.Ghamari, A. et al. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev. 27, 767–777 (2013).
- Show contextfor reference 48
- 49.Iborra, F. J., Pombo, A., Jackson, D. A. & Cook, P. R. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J. Cell Sci. 109(Pt 6), 1427–1436 (1996).
- Show contextfor reference 49
- 50.Meier, M. et al. Cohesin facilitates zygotic genome activation in zebrafish. Development 145, 145 (2018). pii: dev156521.
- Show contextfor reference 50
- 51.Bartfai, R. et al. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr. Biol. 14, 593–598 (2004).
- Show contextfor reference 51
- 52.Garcia-Muse, T. & Aguilera, A. Transcription-replication conflicts: how they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 17, 553–563 (2016).
- Show contextfor reference 52
- 53.Chen, H., Xu, Z., Mei, C., Yu, D. & Small, S. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149, 618–629 (2012).
- Show contextfor reference 53
- 54.Lagha, M. et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987 (2013).
- Show contextfor reference 54
- 55.Hug, C. B., Grimaldi, A. G., Kruse, K. & Vaquerizas, J. M. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 169, 216–228 e19 (2017).
- Show contextfor reference 55
- 56.Blythe, S. A. & Wieschaus, E. F. Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. Elife 5, e20148 (2016).
- Show contextfor reference 56
- 57.Stevens, T. J. et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 544, 59–64 (2017).
- Show contextfor reference 57
- 58.Branco, M. R. & Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4, e138 (2006).
- Show contextfor reference 58
- 59.Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
- Show contextfor reference 59
- 60.Haberle, V., Forrest, A. R. R., Hayashizaki, Y., Carninci, P. & Lenhard, B. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res.43, e51–e51 (2015).
- Show contextfor reference 60
- 61.Kent, W. J. BLAT—The BLAST-Like Alignment Tool. Genome Res.12, 656–664 (2002).
- Show contextfor reference 61
- 62.Sievers, F. & Higgins, D. G. Clustal Omega, Accurate Alignment of Very Large Numbers of Sequences. in Multiple Sequence Alignment Methods (ed. Russell, D. J.) 105-116 (Humana Press, Totowa, NJ, 2014).
- Show contextfor reference 62
- 63.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
- Show contextfor reference 63
- 64.Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
- Show contextfor reference 64
- 65.Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
- Show contextfor reference 65
- 66.Kestler, H. A. et al. VennMaster: Area-proportional Euler diagrams for functional GO analysis of microarrays. BMC Bioinforma. 9, 67 (2008).
- Show contextfor reference 66
- 67.Chen, H. & Boutros, P. C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinforma. 12, 35 (2011).
- Show contextfor reference 67
- 68.Gréen, A., Lönn, A., Peterson, K. H., Öllinger, K. & Rundquist, I. Translocation of histone H1 subtypes between chromatin and cytoplasm during mitosis in normal human fibroblasts. Cytom. Part A 77, 478–484 (2010).
- Show contextfor reference 68
- 69.Mora-Bermúdez, F., Gerlich, D. & Ellenberg, J. Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 9, 822–831 (2007).
- Show contextfor reference 69
- 70.Percival, S. M. et al. Variations in dysfunction of sister chromatid cohesion in esco2 mutant zebrafish reflect the phenotypic diversity of Roberts syndrome. Dis. Models & Mech. 8, 941–955 (2015).
- Show contextfor reference 70
Acknowledgements
This work was supported by a Wellcome Trust Investigator award (106955) and BBSRC project grant to F.M., NEURAM (712821) and VISGEN (734862) projects of H2020 by the European Commission to F.M., A.S., H.K.Q., H.v.N. & J.S.F. BBSRC MIBTP doctoral training programme to L.C., and European Research Council Advanced grant (ERC-2013-340551, Birtoaction) to L.T. We thank Jon Moulton and Yongfu Li at Gene-Tools for advice on morpholino designs and for offering morpholino reagents, the COMPARE imaging facility at the University of Birmingham, Aditi Kanhere and Anja Baranasic for advice in computational analyses, Sarah Baxendale for providing transgenic lines and Phil Zegerman for chk1 and replication factor expression vectors, Marissa Williams and Glen Reid for assistance with the ddPCR and Piotr Balwierz for comments on the manuscript.
Author information
Affiliations
- Institute of Cancer and Genomic Sciences, College of Medical and Dental Sciences, University of Birmingham, Birmingham, B15 2TT, UK
- Yavor Hadzhiev
- , Haseeb K. Qureshi
- , Lucy Wheatley
- , Ledean Cooper
- , Aleksandra Jasiulewicz
- , Joseph W. Wragg
- , Divyasree Poovathumkadavil
- , Agnieszka Gambus
- & Ferenc Müller
- School of Chemistry, University of Birmingham, Edgbaston, Birmingham, B15 2TT, UK
- Huy Van Nguyen
- & John S. Fossey
- Institut de Génétique et de Biologie Moléculaire et Cellulaire, UMR7104, Centre National de la Recherche Scientifique, Strasbourg, 67404, Illkirch, France
- Sascha Conic
- & Làszlò Tora
- Institut National de la Santé et de la Recherche Médicale, U964, Université de Strasbourg, Strasbourg, 67404, Illkirch, France
- Sascha Conic
- & Làszlò Tora
- Faculty of Engineering and Information Technology, Biomedical Engineering School, University of Technology, Sydney, NSW 2007, Australia
- Sarah Bajan
- & György Hutvàgner
- Institute of Transdisciplinary Discoveries, Institute of Physiology, Medical School, University of Pecs, Pecs, H-7624, Hungary
- Attila Sik
- Institute of Clinical Science, College of Medical and Dental Sciences, University of Birmingham, Birmingham, B15 2TT, UK
- Attila Sik
Contributions
F.M., Y.H. conceived and designed the study and wrote the ms with contributions from all other authors, Y.H. designed and carried out most MO injections and imaging experiments, H.K.Q. designed and undertook imaging data analysis, L.W. designed and carried out DNA FISH and immunostaining experiments, L.C. annotated miR430promoters and znf genes, A.J. carried out cell cycle experiments and analyses, H.v.N. and J.S.F. designed and facilitated chemical synthesis of MOs, J.W.W. measured ploidy, S.B. and G.H. designed and carried out measurements of miR430 levels, D.P. and A.G. generated fluorescently labelled proteins, L.T. and S.C. labelled Fab-s against Ser2p of Pol II for in vivo studies and A.S. provided input on experimental designs.
Competing interests
The authors declare no competing interests.
Corresponding author
Correspondence to Ferenc Müller.

